IN 2015, the first commercial nanopore DNA sequencing device was introduced by Oxford Nanopore Technologies. Based on a synthetically engineered transmembrane protein, nanopore sequencing allows long DNA strands to be channelled through the central lumen of the pore where changes in the ionic current work as a sensor of the individual bases in the DNA. This technique was a key milestone for DNA sequencing and the achievement was only made possible after decades of research.
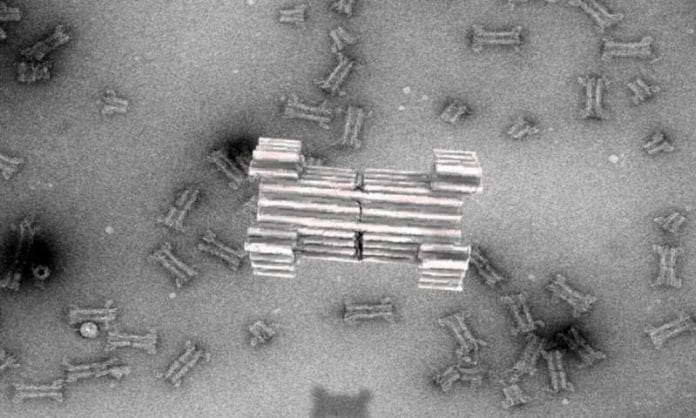
Since then, researchers have tried to extend this principle and build larger pores to accommodate proteins for sensing purposes, but a major challenge has been the limited understanding of artificial protein design. As an alternative, a new technique based on artificial folding of DNA into complex structures, the so-called 3D-origami technique, first reported by the AU group in 2009, has emerged. Compared to proteins, DNA origami has been shown to have an unprecedented design space for constructing nanostructures that mimic and extend naturally occurring complexes.
In a new article, published in Nature Communications, the researchers now report the creation of a large synthetic nanopore made from DNA. This nanopore structure is capable of translocating large protein-sized macromolecules between compartments separated by a lipid bilayer. In addition, a functional gating system was introduced inside the pore to enable biosensing of very few molecules in solution.
With the use of powerful optical microscopes, the researchers could follow the flow of molecules through individual nanopores. By introducing a controllable plug in the pore, it was furthermore possible to size-selectively control the flow of proteinsize molecules and demonstrate label-free, real time, bio-sensing of a trigger molecule.